Pub figures
Some of figures from different segments of the analysis were colated for the manuscript.
Here we present the code/process used to put them together.
Treatments analysis
Number of treatments and proportion of the total dose used in fungicide trials.
Load packages
list.of.packages <-
c(
"tidyverse",
"readxl",
"devtools",
"ggpubr",
"zoo",
"kableExtra",
"conflicted",
"here",
"lubridate",
"magick",
"egg",
"conflicted",
"poppr",
"magick"
)
new.packages <-
list.of.packages[!(list.of.packages %in% installed.packages()[, "Package"])]
#Download packages that are not already present in the library
if (length(new.packages))
install.packages(new.packages)
packages_load <-
lapply(list.of.packages, require, character.only = TRUE)
#Print warning if there is a problem with installing/loading some of packages
if (any(as.numeric(packages_load) == 0)) {
warning(paste("Package/s: ", paste(list.of.packages[packages_load != TRUE], sep = ", "), "not loaded!"))
} else {
print("All packages were successfully loaded.")
}## [1] "All packages were successfully loaded."rm(list.of.packages, new.packages, packages_load)
conflict_prefer("filter", "dplyr")
#if instal is not working try
#install.packages("ROCR", repos = c(CRAN="https://cran.r-project.org/"))Weather
df <-
read_csv(
here::here("data", "weather" , "hly375.csv"),
col_types = cols(
clamt = col_skip(),
clht = col_skip(),
vis = col_skip(),
w = col_skip(),
ww = col_skip()
),
skip = 17
)
df$date <-
lubridate::dmy_hm(df$date)
df<- add_column(df, short_date = as.Date(df$date), .after=1)
df<- add_column(df, year = year(df$date), .after=2)
df<- add_column(df, month = month(df$date), .after=3)
df<- add_column(df, day = day(df$date), .after=4)
infil_gap <- 8 #Maximum length of the infill gap
df$temp <-
round(na.spline(df$temp, na.rm = FALSE, maxgap = infil_gap), 1)
df$rhum <-
round(na.spline(df$rhum, na.rm = FALSE, maxgap = infil_gap), 0)
df$rhum <- sapply(df$rhum, function(x)
ifelse(x > 100, x <- 100, x))
df <-
df %>%
filter(year %in% 2016:2019)%>%
filter(month %in% c(4:9))
#Check if the imputation worked
df %>%
summarize(
NA_rain = sum(is.na(rain)),
NA_temp = sum(is.na(temp)),
NA_rhum = sum(is.na(rhum))
)%>%
kable(format = "html") %>%
kableExtra::kable_styling(latex_options = "striped",full_width = FALSE)| NA_rain | NA_temp | NA_rhum |
|---|---|---|
| 2 | 0 | 0 |
#Only two missing values for rain should not be a problem, so we will just do a linear interpolation
df$rain <-
round(na.approx(df$rain, na.rm = FALSE, maxgap = infil_gap), 1)
dfday <-
df %>%
group_by(year, month, short_date) %>%
summarize(
mintemp = round(min(temp), 1),
minrhum = round(min(rhum), 1),
temp = round(mean(temp), 1),
rain = sum(rain),
rhum = round(mean(rhum), 1),
meantemp = round(mean(temp), 1),
meanrhum = round(mean(rhum), 1)
)
#REad disease data
disdata <-
read_rds( path = here::here("results", "dis", "Duration of the epidemics in years.Rdata"))
colnames(disdata) <-
c("year", "disout", "disend")
dfday <-
left_join(dfday, disdata, by = "year")
# rolling mean
rlmean <- 10
dfday$wrhum <- zoo::rollmean(dfday$meanrhum, rlmean, mean, align = 'center', fill = NA)
dfday$wtemp <- zoo::rollmean(dfday$mintemp, rlmean, mean, align = 'center', fill = NA)
#conducive days
templine <- 10
dfday <-
dfday %>%
mutate(condday = ifelse( wtemp>=templine&rain>=.2&rhum>=80& short_date<disout, 50,NA))
#sum of conditions prior to outbreak
dfday <-
dfday %>%
group_by(year) %>%
summarise(consum= sum(condday/50, na.rm = TRUE )) %>%
left_join(dfday,., by = "year")
#LAbels for cumulative sum of critical days
dfday <-
dfday %>%
mutate(symptoms = paste0("Symptoms (", consum, " cond. days)"))
linesize <- .35 #size of the line for variables
dfday %>%
mutate(rain = ifelse(rain == 0, NA, rain)) %>%
rename(date = short_date) %>%
ggplot() +
geom_point(aes(disout, 46),shape =2, size = 1.5, fill ="black")+
geom_point(aes(disend, 46),shape =2, size = 1.5, fill ="black")+
geom_point(aes(date, condday), shape = 3,size = .55)+
geom_line(
aes(
x = date,
y = rhum,
colour = "Relative humidity (%)"
),
size = linesize
) +
geom_line(aes(
x = date,
y = temp,
colour = "Temperature (˚C)",
),
size = linesize) +
geom_line(aes(
x = date,
y = wtemp,
colour = "Rolling mean temp. (˚C)"
),
linetype = "dashed",
size = linesize) +
geom_col(
aes(date,
rain,
fill = "Total precipitation (mm/day)"),
size = 1.2 ,
inherit.aes = TRUE,
width = 0.8
) +
scale_colour_manual(
"Daily weather:",
values = c(
"Relative humidity (%)" = "#0EBFE9",
"Temperature (˚C)" = "#ED2939",
"Rolling mean temp. (˚C)"= "darkred"
)
) +
scale_fill_manual(name= NA, values = c("Total precipitation (mm/day)" = "blue")
) +
scale_y_continuous( breaks = seq(0,100,10), limits = c(-1,100),expand = c(0, 0))+
scale_x_date(date_labels = "%b", date_breaks = "1 month",expand = c(.03, .04))+
facet_wrap( ~ year, scales = "free", ncol = 1,strip.position="right") +
guides(fill=guide_legend(nrow=1,byrow=TRUE,title.position = "top"),
color=guide_legend(nrow=1,byrow=TRUE,title.position = "top"),
linetype=guide_legend(nrow=1,byrow=TRUE,title.position = "top"))+
theme_bw()+
theme(
text = element_text(size=10.8),
# axis.text.x = element_blank(),
# axis.ticks.x = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
# axis.text.y = element_blank(),
# axis.ticks.y = element_blank(),
legend.position = "top",
strip.background =element_rect(colour = "black",size=0.75),
strip.placement = "outside",
legend.title = element_blank(),
panel.grid.minor = element_blank(),
panel.grid.major = element_line(colour = "lightgray",size=linesize),
plot.margin = margin(0, 0, 0, 0, "cm"),
panel.spacing.y=unit(0, "lines"),
legend.box.spacing = unit(0.1, 'cm'),
legend.margin = margin(0, 0, 0, 0, "cm"),
)+
geom_text(aes(label= symptoms, y = 39, x = disout),size = 2.5)+
geom_text(aes(label= "Last assesment", y = 39, x = disend-1.5),size = 2.5)+
ggsave(filename= here::here("results", "wth", "all_wth_dis.png"),
width = 6.5, height =8, dpi = 720)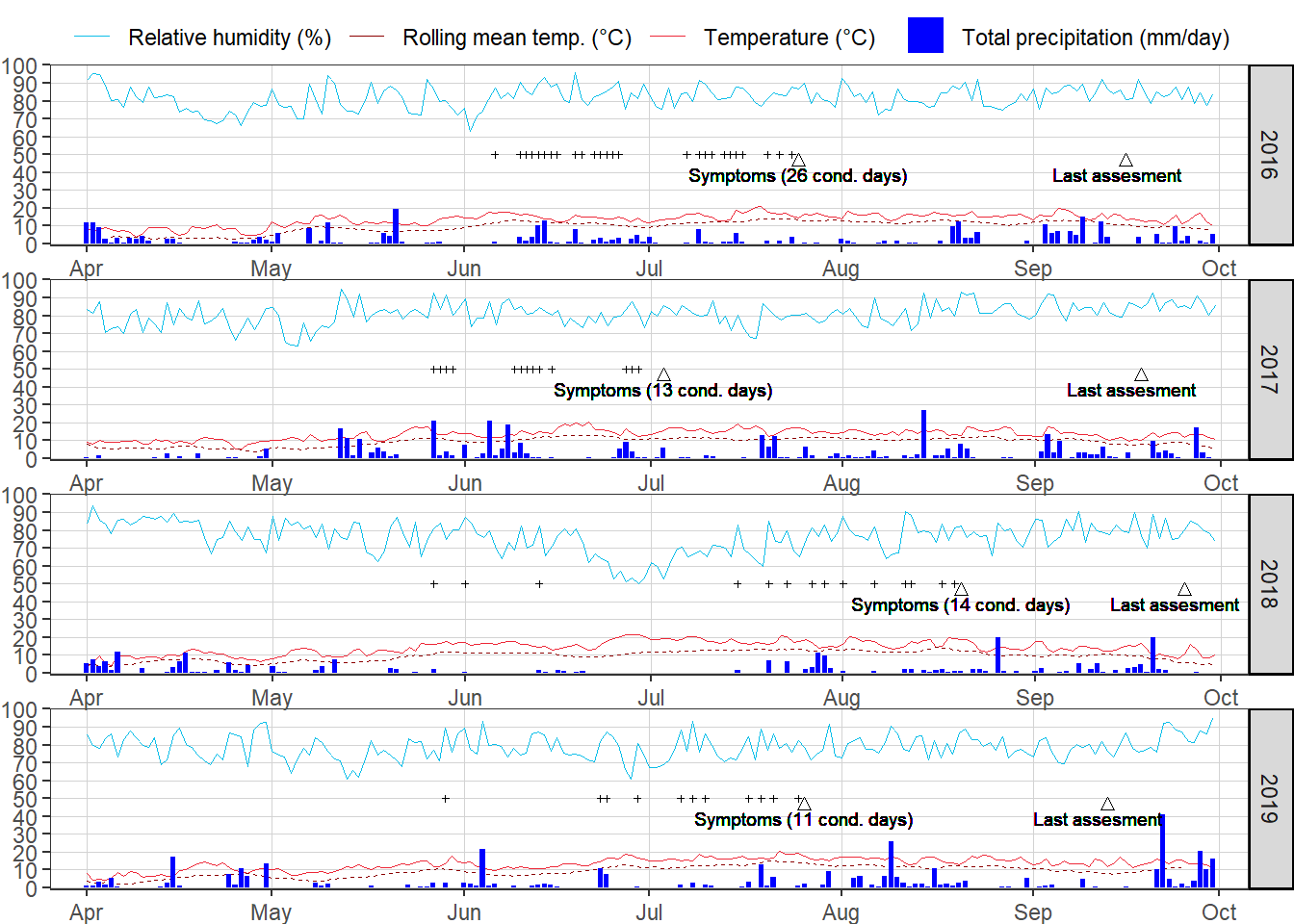
Population frequencies and DAPC analysis
GGplots from the frequency analysis wee colated using ‘ggpubr’ and DAPC plots using ‘magick’.
plotls <- readRDS(file = here::here("results", "gen", "freq","freq_plots.RDS"))
plotls <-
lapply(plotls, function(x)
x <- x+
theme(plot.background = element_rect(size=.3,linetype="solid",color="black")))
plotf <-
ggpubr::ggarrange(plotlist = plotls,
heights = c(1,1,1),
# labels = c("r","rr"),
nrow = 3)
ggsave(
plot = plotf,
filename = here::here("results", "gen", "freq", "Freq_all.png"),
width = 2.9,
height = 9,
dpi = 820
)
# Read external images
#shell.exec(here::here("results", "gen","dapc", "treatment.png"))
t = image_read(here::here("results", "gen","dapc", "treatment.png"))
v = image_read(here::here("results", "gen","dapc", "variety.png"))
y = image_read(here::here("results", "gen","dapc", "year.png"))
imgs = c(v,t,y)
# concatenate them left-to-right (use 'stack=T' to do it top-to-bottom)
side_by_side = image_append(imgs, stack=T)
# save
image_write(side_by_side,
path = here::here("results", "gen","dapc","dapcs", "full_dapc.png"),
format = "png")
#add the frequency plots
f = image_read(here::here("results", "gen", "freq", "Freq_all.png"))
dapcs = image_read(here::here("results", "gen","dapc","dapcs", "full_dapc.png"))
#Append and save the image
scale <- paste0("x", 7380)
# concatenate them left-to-right (use 'stack=T' to do it top-to-bottom)
side_by_side <- image_append(c(f, image_scale(dapcs, scale)), stack = FALSE)
image_write(side_by_side,
path = here::here("results", "gen","dapc","dapcs", paste0(scale, "full.png")),
format = "png",
# width = 4500,
# height = 3700
)
#To open the figure in external viewer uncoment the following lines
#shell.exec(here::here("results", "gen","dapc","dapcs", paste0(scale, "full.png")))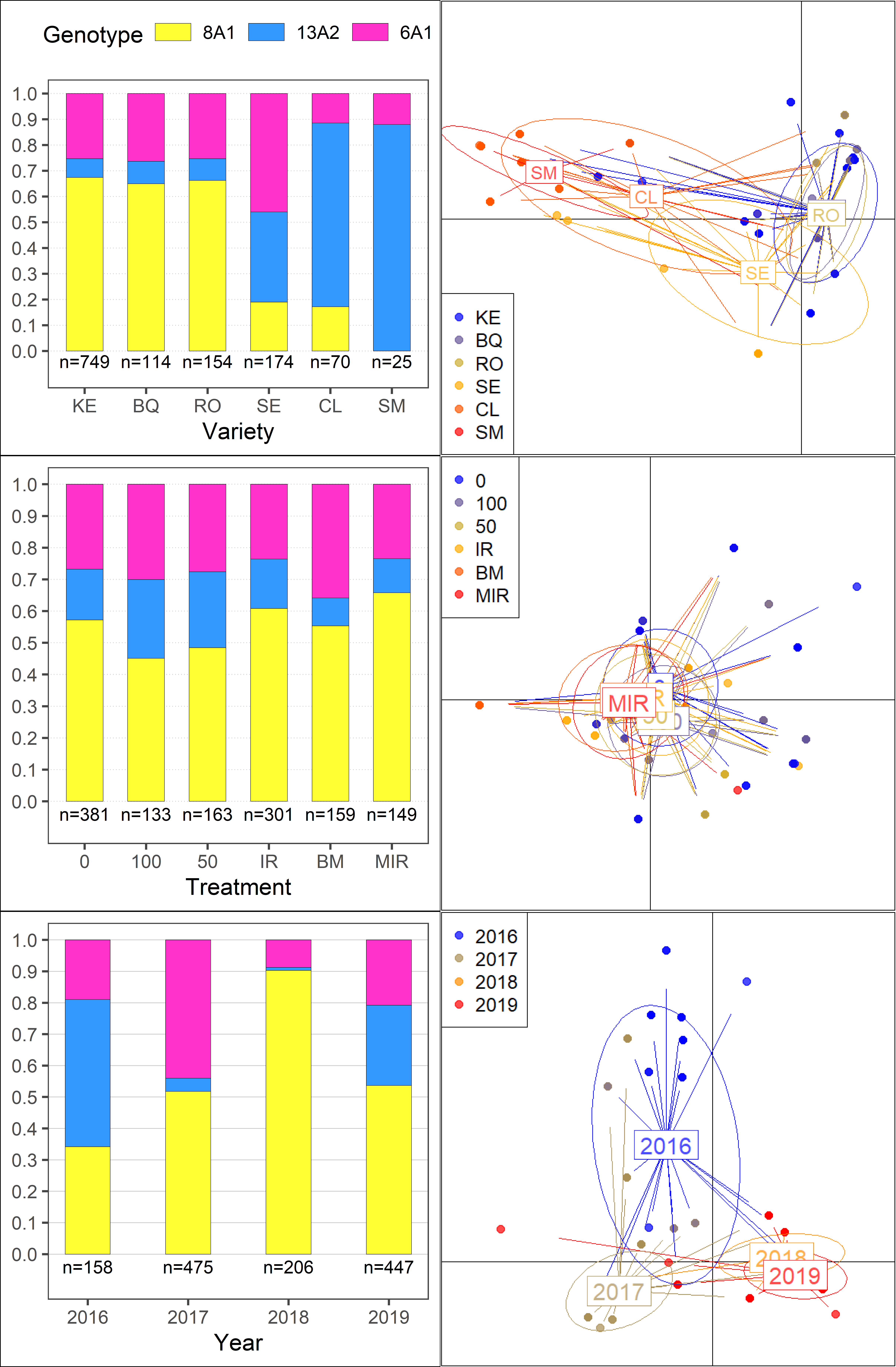
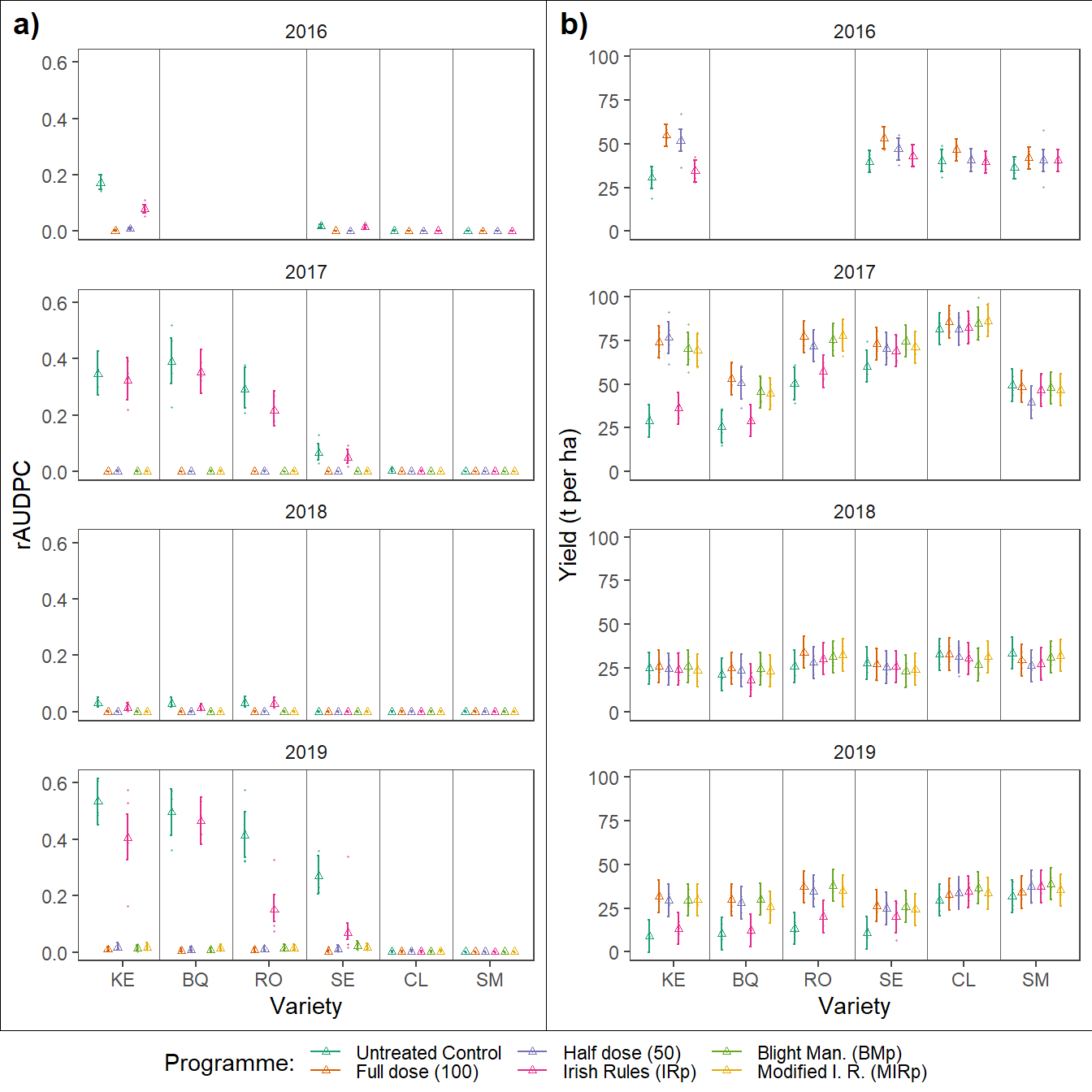
Final figure
Disease and yield
Results of the foliar disease controll yield analysis were colated in single figure using ‘magick’ package.
dd <-
readRDS( file = here::here("results", "dis", "dis_fin.RDS"))
yd <-
readRDS( file = here::here("results", "yield", "yld_fin.RDS"))
yld <-
readRDS( file = here::here("results", "yield", "yld_dat_fin.RDS"))
audpc_data <-
readRDS( file = here::here("results", "dis", "dis_audpc_fin.RDS"))
dodging <- .8
pdfin <-
dd %>%
mutate(
line_positions = as.numeric(factor(variety, levels = unique(variety))),
line_positions = line_positions + .5,
line_positions = ifelse(line_positions == max(line_positions), NA, line_positions),
line_positions = ifelse(year == 2016 &
variety == "SM", 5.5, line_positions),
line_positions = ifelse(year == 2016 &
variety == "CL", 4.5, line_positions),
line_positions = ifelse(year == 2016 &
variety == "SE", 3.5, line_positions),
line_positions = ifelse(year == 2016 &
variety == "KE", 1.5, line_positions)
) %>%
ggplot(data = ., aes(x = variety, y = prop)) +
geom_errorbar(
aes(
ymin = lower.CL,
ymax = upper.CL,
group = treatment,
color = treatment
),
position = position_dodge(width = dodging),
width = .2
) +
geom_point(
aes(y = prop, group = treatment, color = treatment),
size = 1,
shape = 2,
position = position_dodge(width = dodging)
)+
geom_point(
data = subset(audpc_data),
aes(y = rAUDPC_adj, color = treatment, group = treatment),
size = .2,
alpha = .5,
position = position_dodge(width = dodging)
) +
facet_wrap(~ year, ncol = 1)+
scale_color_brewer("Programme:", palette = "Dark2") +
theme_article() +
theme(legend.position = "bottom",
text = element_text(size = 13)) +
geom_vline(aes(xintercept = line_positions),
size = .2,
alpha = .6) +
labs(colour = "Programme:",
x = "Variety",
y = "rAUDPC")
ggsave(plot = pdfin,
filename = here::here("results", "dis", "Effects final ver.png"),
width = 4,
height = 7,
dpi = 620
)
py_fin <-
yd %>%
mutate(
line_positions = as.numeric(factor(variety, levels = unique(variety))),
line_positions = line_positions + .5,
line_positions = ifelse(line_positions == max(line_positions), NA, line_positions),
line_positions = ifelse(year == 2016 &
variety == "SM", 5.5, line_positions),
line_positions = ifelse(year == 2016 &
variety == "SE", 4.5, line_positions)
) %>%
ggplot(data = ., aes(x = variety, y = lsmean)) +
geom_errorbar(
aes(
ymin = lower.CL,
ymax = upper.CL,
group = treatment,
color = treatment
),
position = position_dodge(width = dodging),
width = .2
) +
geom_point(
aes(y = lsmean, group = treatment, color = treatment),
size = 1,
shape = 2,
position = position_dodge(width = dodging)
) +
geom_point(
data = yld,
aes(y = marketable, color = treatment, group = treatment),
size = .2,
alpha = .5,
position = position_dodge(width = dodging)
) +
facet_wrap(~ year, ncol = 1) +
scale_color_brewer("Programme:", palette = "Dark2") +
theme_article() +
theme(legend.position = "bottom",
text = element_text(size = 13)) +
geom_vline(aes(xintercept = line_positions),
size = .2,
alpha = .6) +
labs(colour = "Programme:",
x = "Variety",
y = "Yield (t per ha)")
py_fin <-
py_fin+
facet_wrap(~ year, ncol = 1)
ggsave(
py_fin,
filename = here::here("results", "yield", "Effects final ver.png"),
width = 3.5,
height = 7,
dpi = 620
)
plotls <- list(pdfin,py_fin)
plotls <-
lapply(plotls, function(x)
x <- x+
theme(plot.background = element_rect(size=.3,linetype="solid",color="black"),
text = element_text(size= 11))
)
ggpubr::ggarrange(plotlist = plotls,
widths = c(2.5,2.5),
heights = c(.5,.5),
labels = c("a)","b)"),
nrow = 1,
common.legend = TRUE,
legend = "bottom")+
ggsave(filename= here::here("results", "dis", "dis_yld.png"),
width = 7.5, height =10, dpi = 620)