Pop
Popoulation analysis
After gaining insight into population structure we can move onto the analysis of
Load packages
list.of.packages <-
c(
"tidyverse",
"devtools",
"here",
"readxl",
"poppr",
"egg",
"conflicted"
)
new.packages <-
list.of.packages[!(list.of.packages %in% installed.packages()[, "Package"])]
#Download packages that are not already present in the library
if (length(new.packages))
install.packages(new.packages)
packages_load <-
lapply(list.of.packages, require, character.only = TRUE)
#Print warning if there is a problem with installing/loading some of packages
if (any(as.numeric(packages_load) == 0)) {
warning(paste("Package/s: ", paste(list.of.packages[packages_load != TRUE], sep = ", "), "not loaded!"))
} else {
print("All packages were successfully loaded.")
}## [1] "All packages were successfully loaded."rm(list.of.packages, new.packages, packages_load)
#if instal is not working try
#install.packages("package_name", repos = c(CRAN="https://cran.r-project.org/"))Data import/preparation
fin <- readRDS(file = here::here("data", "gen data", "final", "gendata.rds") )Prepare the data in genalex format.
fin <-
unite(fin, "Ind", idd, year, remove = F)
fin <-
unite(fin, "Pop", variety, Genotype, year)
popcol <- match("Pop",names(fin))
idcol <- match("Ind",names(fin))
gen <-
fin[ , c(idcol,popcol,match("D13",names(fin)):match("X__24",names(fin)))]
path <- here::here("data", "gen data", "final", "pop.csv")
data.frame(ssr = c(12,"PInf_Ireland"),
samples = nrow(gen),
pop = length(unique(fin$Pop)))write.table(data.frame(ssr = 12, samples = nrow(gen), pop = length(unique(fin$Pop))),
path,
sep=",", col.names=FALSE, row.names = FALSE)
write.table( "PInf_Ireland",
path,
sep = ",", row.names = FALSE,
col.names = FALSE, append = T)
names(gen)[grep("X__", names(gen))] <- ""
write.table(gen,
path,
sep = ",", row.names = FALSE,
col.names = !file.exists("myDF.csv"), append = T)Bruvo charts
path <- here::here("data", "gen data", "final", "pop.csv")
monpop <- read.genalex(path, ploidy = 3)
splitStrata(monpop) <- ~variety/genotype/year
monpop##
## This is a genclone object
## -------------------------
## Genotype information:
##
## 29 original multilocus genotypes
## 1287 triploid individuals
## 12 codominant loci
##
## Population information:
##
## 3 strata - variety, genotype, year
## 52 populations defined -
## KE_6A1_2016, KE_8A1_2016, SE_6A1_2016, ..., KE_36A2_2019, SM_13A2_2019, SM_6A1_2019monpop$strata$genotype <- factor( monpop$strata$genotype, levels = c( "8A1","13A2","6A1", "36A2"))
# imsn()
monpop_sub <- popsub(monpop, blacklist = character(0))
monpop_dist <- diss.dist(monpop_sub, percent = FALSE, mat = FALSE)
min_span_net <- poppr.msn(monpop_sub, monpop_dist, showplot = FALSE, include.ties = TRUE)
#Bruvo distance Genotype
setPop(monpop) <- ~genotype
monpop_sub <- popsub(monpop, blacklist = character(0))
min_span_net <- bruvo.msn(monpop_sub, replen = c(1, 2, 3), add = TRUE, loss = TRUE, showplot = FALSE, include.ties = TRUE)
set.seed(1000)
cbbPalette <- c ( "#ff33cc", # pink 6
"#ffff33", # yellow 8
"#3399ff",#blue 13
"#ffcccc"
)Bruvo genotypes
min_span_net <-
bruvo.msn(
monpop,
replen = c(1, 2, 3),
add = TRUE,
loss = TRUE,
showplot = FALSE,
include.ties = TRUE
)
plot_poppr_msn(monpop,
min_span_net,
mlg = FALSE,
gadj = 7,
nodescale = 4,
nodelab = 1,
palette =cbbPalette,
cutoff = 11,
inds = "none",
quantiles = FALSE,
beforecut = TRUE,
pop.leg = TRUE,
size.leg = TRUE,
scale.leg = TRUE,
layfun = igraph::layout_nicely)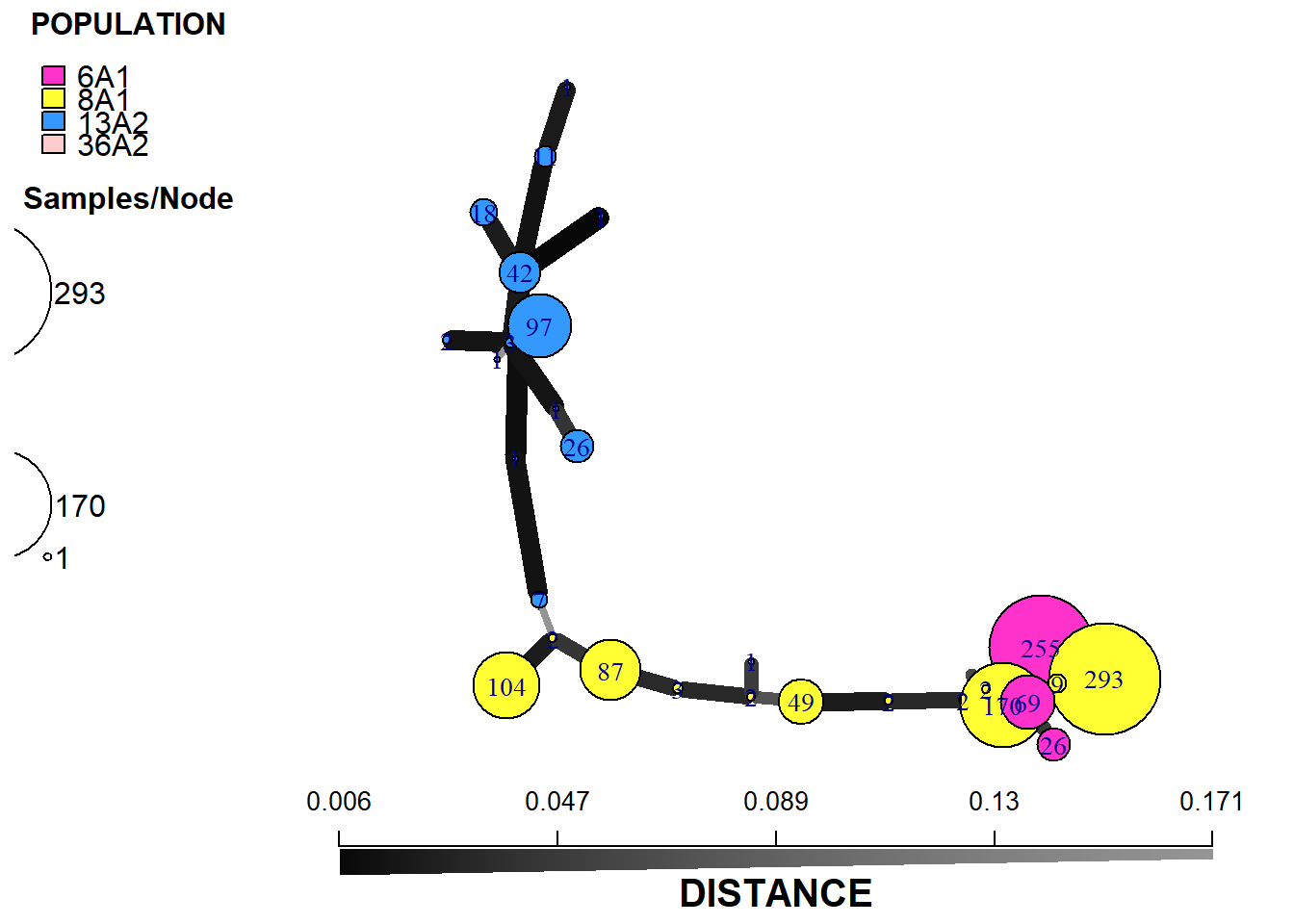
dev.copy(png,filename=here::here("results", "gen","bruvo", "Pop.png"),
width = 700, height= 700 );## png
## 3dev.off ()## png
## 2Bruvo year
setPop(monpop) <- ~year
monpop_sub <- popsub(monpop, blacklist = character(0))
min_span_net <- bruvo.msn(monpop_sub, replen = c(1, 2, 3), add = TRUE, loss = TRUE, showplot = FALSE, include.ties = TRUE)
set.seed(1000)
cbbPalette <- c ("#ff33cc", "#ffff33","#3399ff")
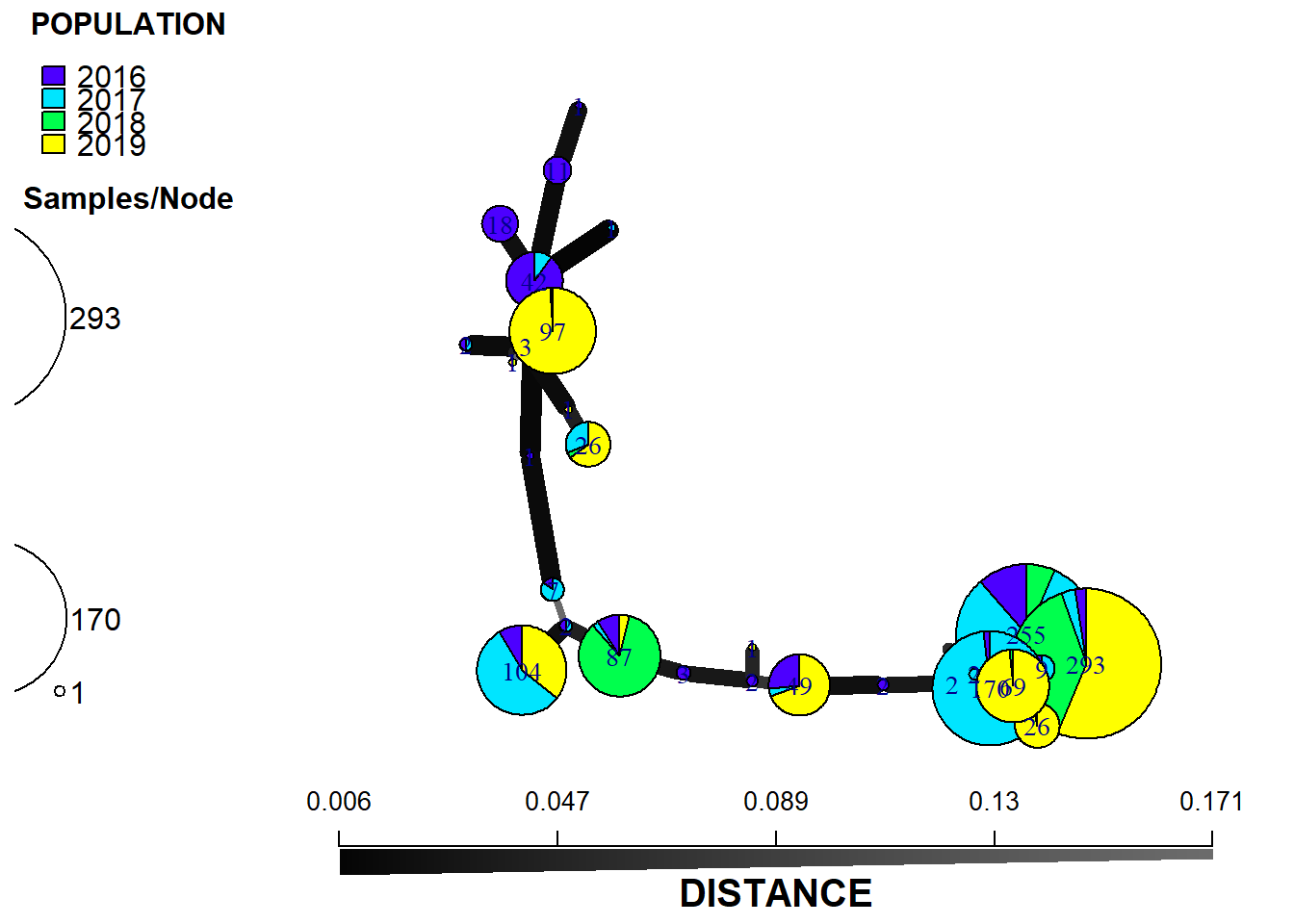
plot_poppr_msn(
monpop,
min_span_net,
mlg = T,
gadj = 4,
inds = "none",
wscale = TRUE,
nodescale = 8,
nodelab = 0.15,
palette = cbbPalette,
cutoff = 0.9,
quantiles = FALSE,
beforecut = TRUE,
layfun = igraph::layout_nicely
)
dev.copy(png,filename=here::here("results", "gen","bruvo", "Year.png"),
width = 1000, height= 700 );## png
## 3dev.off ()## png
## 2Bruvo variety
setPop(monpop) <- ~variety
monpop_sub <- popsub(monpop, blacklist = character(0))
min_span_net <- bruvo.msn(monpop_sub, replen = c(1, 2, 3), add = TRUE, loss = TRUE, showplot = FALSE, include.ties = TRUE)
set.seed(1000)
plot_poppr_msn(monpop,
min_span_net,
inds = "none",
mlg = FALSE,
gadj = 6,
nodescale = 8,
palette = rainbow,
cutoff = 0.2,
quantiles = FALSE,
beforecut = TRUE,
pop.leg = TRUE,
size.leg = TRUE,
scale.leg = TRUE,
layfun = igraph::layout_nicely)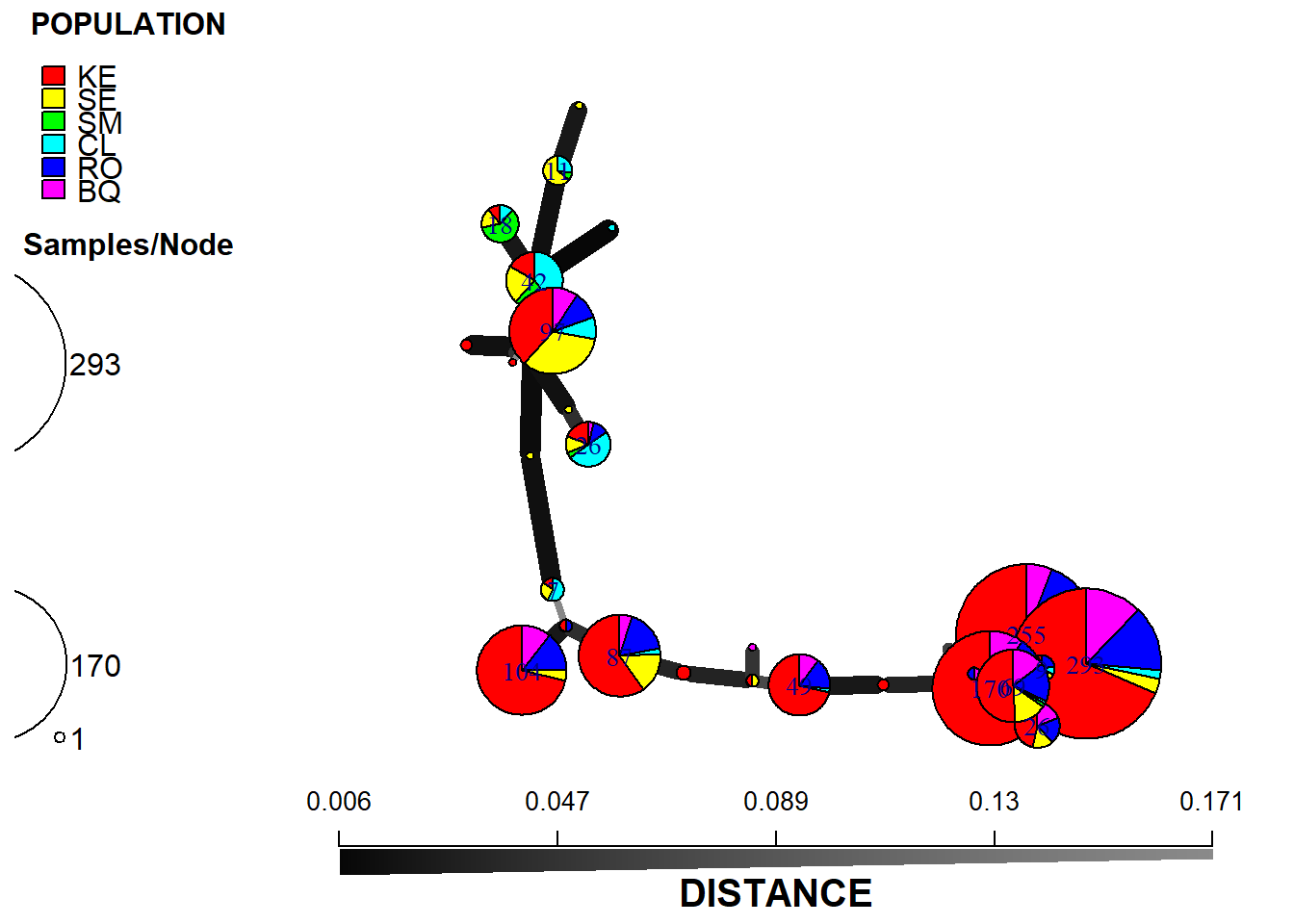
Bruvo year and genotype
setPop(monpop) <- ~year/genotype
monpop_sub <- popsub(monpop, blacklist = character(0))
min_span_net <- bruvo.msn(monpop_sub, replen = c(1, 2, 3), add = TRUE, loss = TRUE, showplot = FALSE, include.ties = TRUE)
set.seed(1000)
plot_poppr_msn(monpop,
min_span_net,
inds = "none",
mlg = FALSE,
gadj = 6,
nodescale = 8,
palette = rainbow,
cutoff = 0.2,
quantiles = FALSE,
beforecut = TRUE,
pop.leg = TRUE,
size.leg = TRUE,
scale.leg = TRUE,
layfun = igraph::layout_nicely)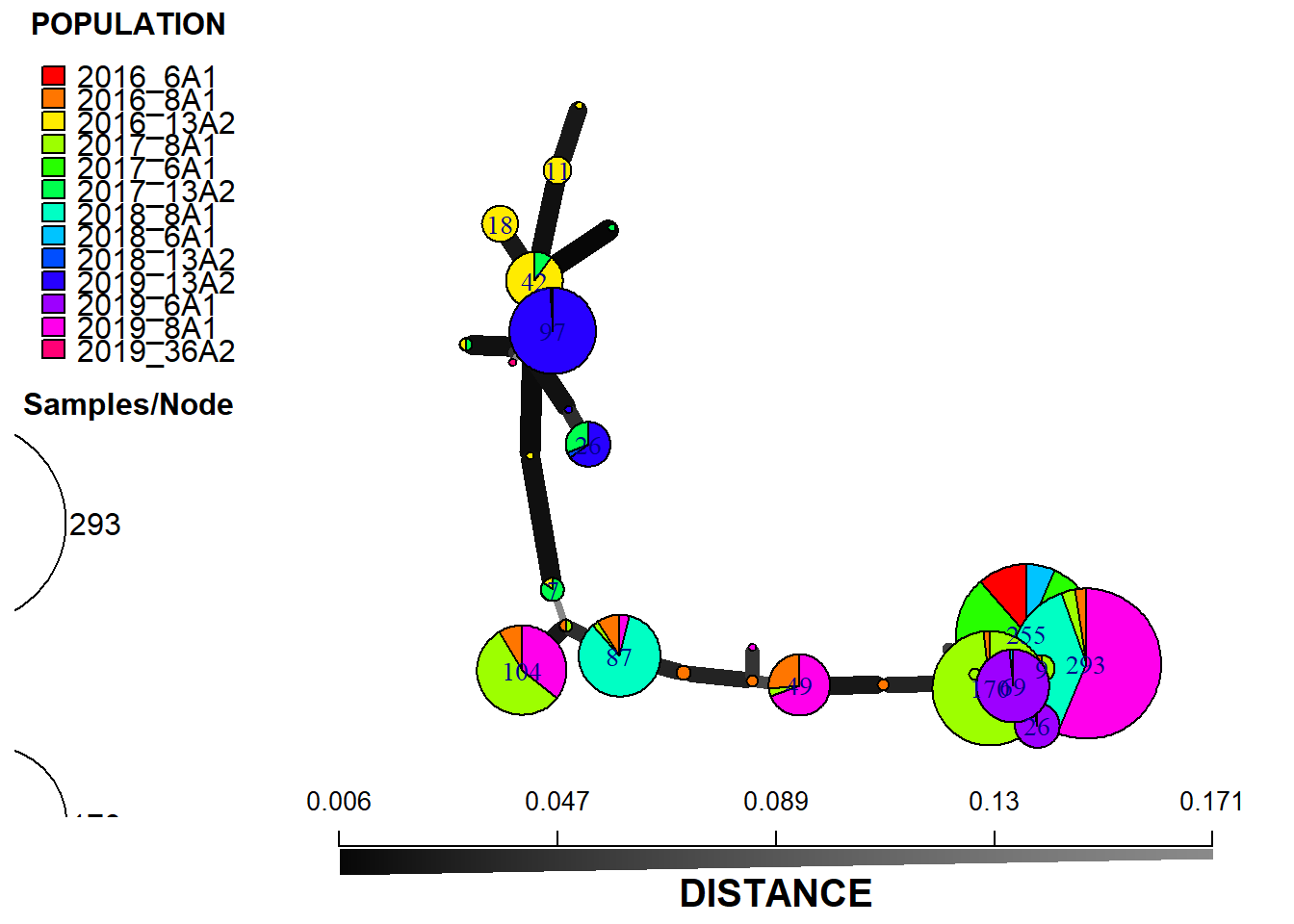
Bruvo year and variety
setPop(monpop) <- ~year/variety
monpop_sub <- popsub(monpop, blacklist = character(0))
min_span_net <- bruvo.msn(monpop_sub, replen = c(1, 2, 3), add = TRUE, loss = TRUE, showplot = FALSE, include.ties = TRUE)
set.seed(1000)
plot_poppr_msn(monpop,
min_span_net,
inds = "none",
mlg = FALSE,
gadj = 6,
nodescale = 8,
palette = rainbow,
cutoff = 0.2,
quantiles = FALSE,
beforecut = TRUE,
pop.leg = TRUE,
size.leg = TRUE,
scale.leg = TRUE,
layfun = igraph::layout_nicely)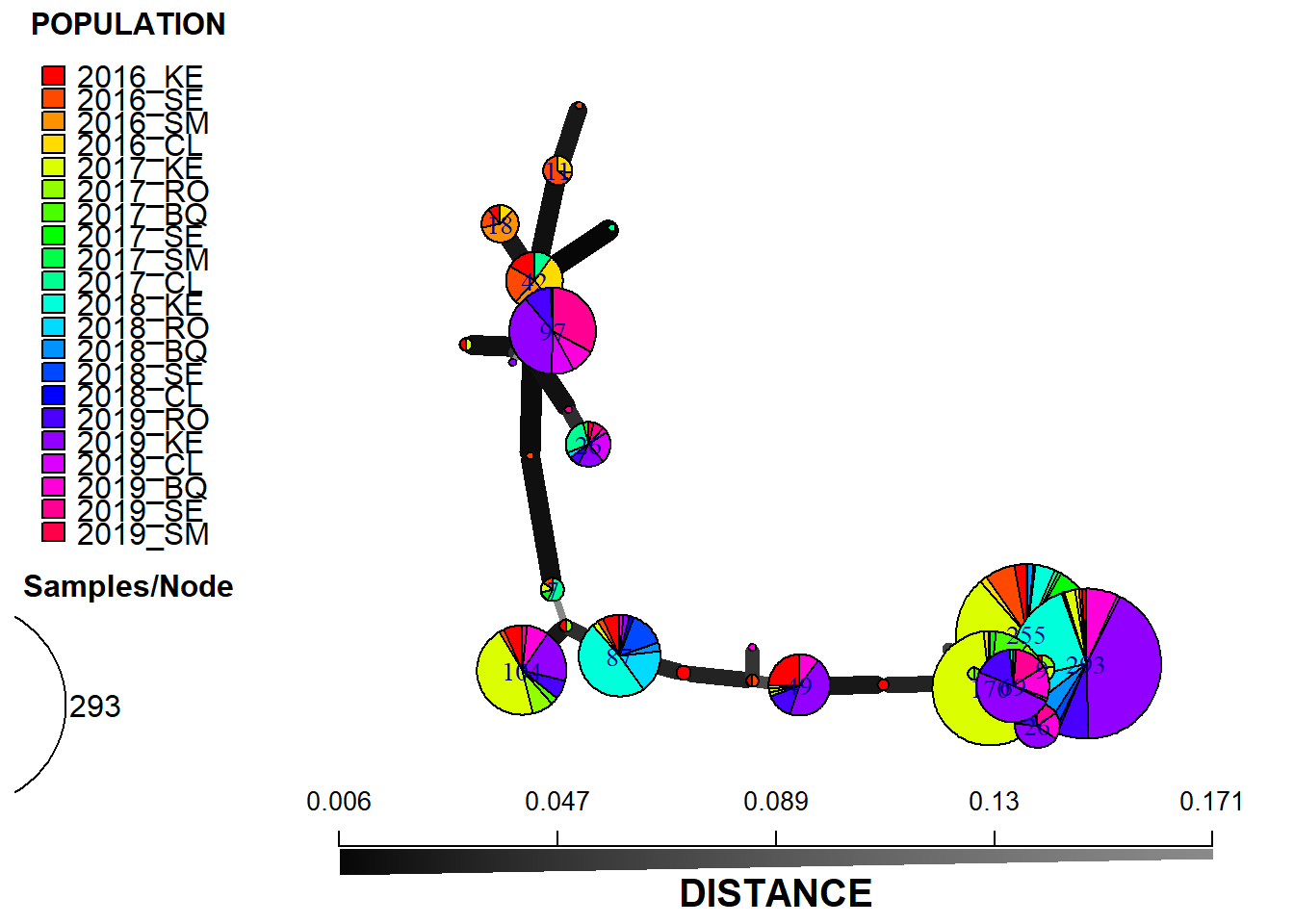
session_info()## - Session info ----------------------------------------------------------
## setting value
## version R version 3.6.1 (2019-07-05)
## os Windows 10 x64
## system x86_64, mingw32
## ui RTerm
## language (EN)
## collate English_United States.1252
## ctype English_United States.1252
## tz Europe/London
## date 2020-03-06
##
## - Packages --------------------------------------------------------------
## package * version date lib source
## ade4 * 1.7-13 2018-08-31 [1] CRAN (R 3.6.1)
## adegenet * 2.1.1 2018-02-02 [1] CRAN (R 3.6.1)
## ape 5.3 2019-03-17 [1] CRAN (R 3.6.1)
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 3.6.1)
## backports 1.1.5 2019-10-02 [1] CRAN (R 3.6.1)
## boot 1.3-22 2019-04-02 [2] CRAN (R 3.6.1)
## broom 0.5.2 2019-04-07 [1] CRAN (R 3.6.1)
## callr 3.3.2 2019-09-22 [1] CRAN (R 3.6.1)
## cellranger 1.1.0 2016-07-27 [1] CRAN (R 3.6.1)
## class 7.3-15 2019-01-01 [2] CRAN (R 3.6.1)
## classInt 0.4-2 2019-10-17 [1] CRAN (R 3.6.1)
## cli 1.1.0 2019-03-19 [1] CRAN (R 3.6.1)
## cluster 2.1.0 2019-06-19 [2] CRAN (R 3.6.1)
## coda 0.19-3 2019-07-05 [1] CRAN (R 3.6.1)
## colorspace 1.4-1 2019-03-18 [1] CRAN (R 3.6.1)
## conflicted * 1.0.4 2019-06-21 [1] CRAN (R 3.6.1)
## crayon 1.3.4 2017-09-16 [1] CRAN (R 3.6.1)
## DBI 1.0.0 2018-05-02 [1] CRAN (R 3.6.1)
## deldir 0.1-23 2019-07-31 [1] CRAN (R 3.6.1)
## desc 1.2.0 2018-05-01 [1] CRAN (R 3.6.1)
## devtools * 2.2.1 2019-09-24 [1] CRAN (R 3.6.1)
## digest 0.6.23 2019-11-23 [1] CRAN (R 3.6.2)
## dplyr * 0.8.3 2019-07-04 [1] CRAN (R 3.6.1)
## e1071 1.7-2 2019-06-05 [1] CRAN (R 3.6.1)
## egg * 0.4.5 2019-07-13 [1] CRAN (R 3.6.1)
## ellipsis 0.3.0 2019-09-20 [1] CRAN (R 3.6.1)
## evaluate 0.14 2019-05-28 [1] CRAN (R 3.6.1)
## expm 0.999-4 2019-03-21 [1] CRAN (R 3.6.1)
## fastmap 1.0.1 2019-10-08 [1] CRAN (R 3.6.1)
## fastmatch 1.1-0 2017-01-28 [1] CRAN (R 3.6.0)
## forcats * 0.4.0 2019-02-17 [1] CRAN (R 3.6.1)
## fs 1.3.1 2019-05-06 [1] CRAN (R 3.6.1)
## gdata 2.18.0 2017-06-06 [1] CRAN (R 3.6.0)
## generics 0.0.2 2018-11-29 [1] CRAN (R 3.6.1)
## ggplot2 * 3.2.1 2019-08-10 [1] CRAN (R 3.6.1)
## glue 1.3.1 2019-03-12 [1] CRAN (R 3.6.1)
## gmodels 2.18.1 2018-06-25 [1] CRAN (R 3.6.1)
## gridExtra * 2.3 2017-09-09 [1] CRAN (R 3.6.1)
## gtable 0.3.0 2019-03-25 [1] CRAN (R 3.6.1)
## gtools 3.8.1 2018-06-26 [1] CRAN (R 3.6.0)
## haven 2.1.1 2019-07-04 [1] CRAN (R 3.6.1)
## here * 0.1 2017-05-28 [1] CRAN (R 3.6.1)
## hms 0.5.2 2019-10-30 [1] CRAN (R 3.6.1)
## htmltools 0.4.0 2019-10-04 [1] CRAN (R 3.6.1)
## httpuv 1.5.2 2019-09-11 [1] CRAN (R 3.6.1)
## httr 1.4.1 2019-08-05 [1] CRAN (R 3.6.1)
## igraph 1.2.4.1 2019-04-22 [1] CRAN (R 3.6.1)
## jsonlite 1.6 2018-12-07 [1] CRAN (R 3.6.1)
## KernSmooth 2.23-15 2015-06-29 [2] CRAN (R 3.6.1)
## knitr 1.25 2019-09-18 [1] CRAN (R 3.6.1)
## later 1.0.0 2019-10-04 [1] CRAN (R 3.6.1)
## lattice 0.20-38 2018-11-04 [2] CRAN (R 3.6.1)
## lazyeval 0.2.2 2019-03-15 [1] CRAN (R 3.6.1)
## LearnBayes 2.15.1 2018-03-18 [1] CRAN (R 3.6.0)
## lifecycle 0.1.0 2019-08-01 [1] CRAN (R 3.6.1)
## lubridate 1.7.4 2018-04-11 [1] CRAN (R 3.6.1)
## magrittr 1.5 2014-11-22 [1] CRAN (R 3.6.1)
## MASS 7.3-51.4 2019-03-31 [2] CRAN (R 3.6.1)
## Matrix 1.2-17 2019-03-22 [2] CRAN (R 3.6.1)
## memoise 1.1.0 2017-04-21 [1] CRAN (R 3.6.1)
## mgcv 1.8-28 2019-03-21 [2] CRAN (R 3.6.1)
## mime 0.7 2019-06-11 [1] CRAN (R 3.6.0)
## modelr 0.1.5 2019-08-08 [1] CRAN (R 3.6.1)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 3.6.1)
## nlme 3.1-140 2019-05-12 [2] CRAN (R 3.6.1)
## pegas 0.12 2019-10-05 [1] CRAN (R 3.6.1)
## permute 0.9-5 2019-03-12 [1] CRAN (R 3.6.1)
## phangorn 2.5.5 2019-06-19 [1] CRAN (R 3.6.1)
## pillar 1.4.2 2019-06-29 [1] CRAN (R 3.6.1)
## pkgbuild 1.0.6 2019-10-09 [1] CRAN (R 3.6.1)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 3.6.1)
## pkgload 1.0.2 2018-10-29 [1] CRAN (R 3.6.1)
## plyr 1.8.4 2016-06-08 [1] CRAN (R 3.6.1)
## polysat 1.7-4 2019-03-06 [1] CRAN (R 3.6.1)
## poppr * 2.8.3 2019-06-18 [1] CRAN (R 3.6.1)
## prettyunits 1.0.2 2015-07-13 [1] CRAN (R 3.6.1)
## processx 3.4.1 2019-07-18 [1] CRAN (R 3.6.1)
## promises 1.1.0 2019-10-04 [1] CRAN (R 3.6.1)
## ps 1.3.0 2018-12-21 [1] CRAN (R 3.6.1)
## purrr * 0.3.3 2019-10-18 [1] CRAN (R 3.6.1)
## quadprog 1.5-7 2019-05-06 [1] CRAN (R 3.6.0)
## R6 2.4.0 2019-02-14 [1] CRAN (R 3.6.1)
## Rcpp 1.0.2 2019-07-25 [1] CRAN (R 3.6.1)
## readr * 1.3.1 2018-12-21 [1] CRAN (R 3.6.1)
## readxl * 1.3.1 2019-03-13 [1] CRAN (R 3.6.1)
## remotes 2.1.0 2019-06-24 [1] CRAN (R 3.6.1)
## reshape2 1.4.3 2017-12-11 [1] CRAN (R 3.6.1)
## rlang 0.4.1 2019-10-24 [1] CRAN (R 3.6.1)
## rmarkdown 1.16 2019-10-01 [1] CRAN (R 3.6.1)
## rprojroot 1.3-2 2018-01-03 [1] CRAN (R 3.6.1)
## rstudioapi 0.10 2019-03-19 [1] CRAN (R 3.6.1)
## rvest 0.3.4 2019-05-15 [1] CRAN (R 3.6.1)
## scales 1.0.0 2018-08-09 [1] CRAN (R 3.6.1)
## seqinr 3.6-1 2019-09-07 [1] CRAN (R 3.6.1)
## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 3.6.1)
## sf 0.8-0 2019-09-17 [1] CRAN (R 3.6.1)
## shiny 1.4.0 2019-10-10 [1] CRAN (R 3.6.1)
## sp 1.3-1 2018-06-05 [1] CRAN (R 3.6.1)
## spData 0.3.2 2019-09-19 [1] CRAN (R 3.6.1)
## spdep 1.1-3 2019-09-18 [1] CRAN (R 3.6.1)
## stringi 1.4.3 2019-03-12 [1] CRAN (R 3.6.0)
## stringr * 1.4.0 2019-02-10 [1] CRAN (R 3.6.1)
## testthat 2.2.1 2019-07-25 [1] CRAN (R 3.6.1)
## tibble * 2.1.3 2019-06-06 [1] CRAN (R 3.6.1)
## tidyr * 1.0.0 2019-09-11 [1] CRAN (R 3.6.1)
## tidyselect 0.2.5 2018-10-11 [1] CRAN (R 3.6.1)
## tidyverse * 1.2.1 2017-11-14 [1] CRAN (R 3.6.1)
## units 0.6-5 2019-10-08 [1] CRAN (R 3.6.1)
## usethis * 1.5.1 2019-07-04 [1] CRAN (R 3.6.1)
## vctrs 0.2.0 2019-07-05 [1] CRAN (R 3.6.1)
## vegan 2.5-6 2019-09-01 [1] CRAN (R 3.6.1)
## withr 2.1.2 2018-03-15 [1] CRAN (R 3.6.1)
## xfun 0.10 2019-10-01 [1] CRAN (R 3.6.1)
## xml2 1.2.2 2019-08-09 [1] CRAN (R 3.6.1)
## xtable 1.8-4 2019-04-21 [1] CRAN (R 3.6.1)
## yaml 2.2.0 2018-07-25 [1] CRAN (R 3.6.0)
## zeallot 0.1.0 2018-01-28 [1] CRAN (R 3.6.1)
##
## [1] C:/Users/mlade/Documents/R/win-library/3.6
## [2] C:/Program Files/R/R-3.6.1/library